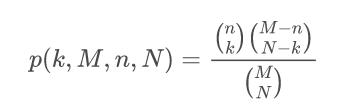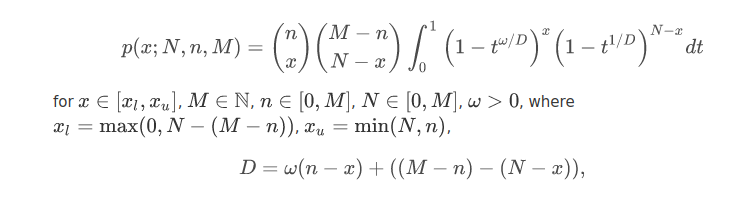1- General info
GeneCodis is an API and web tool for the Singular and Modular Enrichment Analysis (SEA and MEA) of lists of genes, proteins, and regulatory elements: miRNAs, transcription factors (TFs), and CpGs. Look CoAnnotation in the next panel for more information on the Modular Enrichment Analysis. The enrichment analysis of regulatory elements, requires a different approach. Due to a gene selection bias (some genes are more targeted than others) in platforms and regulation networks it requires a test based on a non-central hypergeometric distribution, concretely, we use the Wallenius distribution. Our method is based on the GOseq implementation. Read the paper for more information: Functional Enrichment Analysis of Regulatory Elements (Biomedicines 2022).
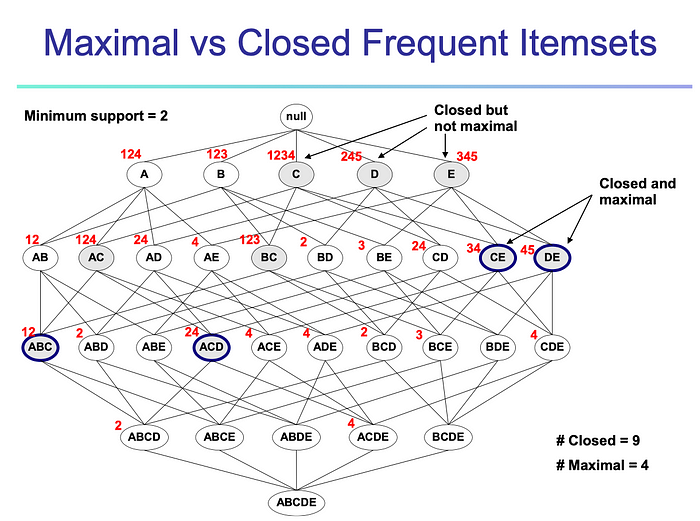
We have updated the annotations and organisms. Annotations are now divided in four general collections, Functional, Regulatory with miRNAs-based annotations, Drugs and Phenotypes. The results are also displayed differently, we are now using D3.js to create two interactive visualizations, a gene-, mirna-annotation network and a bars chart.
In case that you are missing any functionality, organisms, annotation or even if you want some kind of new feature please do not hesitate in contact us at statsandcompbio@outlook.com since we are continually working on creating a complete updated GeneCodis, feedback is always appreciated. We want GeneCodis to be an open source project and share it with all developers and researchers, so feel free to visit our Github repository and fork your own branch.